CS 5043: HW6: Recurrent Neural Networks and Timeseries Data
Assignment notes:
- Deadline:
- Expected completion: Tuesday, April 20th @11:59pm.
- Deadline: Thursday, April 22nd @11:59pm.
- Hand-in procedure: submit a pdf to Gradescope
- This work is to be done on your own. While general discussion
about Python, Keras and Tensorflow is encouraged, sharing
solution-specific code is inappropriate. Likewise, downloading
solution-specific code is not allowed.
- Do not submit MSWord documents.
The Problem
Peptides are short chains of amino acids that perform a wide range of
biological functions, depending on the specific sequence of amino
acids. Understanding how amino acids interact with other molecules is
key to understanding the mechanisms of genetic diseases and to
constructing therapies to address these diseases.
One form of interaction is the question of how well a peptide binds to
a specific molecule (this is known as binding affinity). For
this assignment, we will use a data set that includes a measure of
binding affinity for a large number of peptides toward two molecules,
known as HLA-DRB1*13:01 and HLA-DRB1*15:01. Our goal is to predict
the binding affinity for novel peptides toward these molecules.
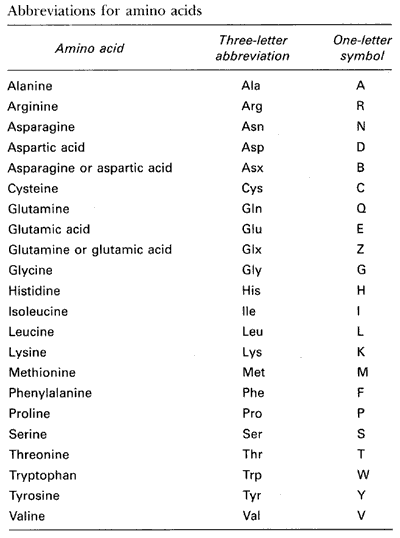
Amino Acids
A total of 22 amino acids make up the building blocks for all peptides
and proteins. The following table gives the names and one-letter
codes used to represent these amino acids:
Data Set
The data
set is available in the git repository in a set of CSV
files. The data are already partitioned into five independent folds
of training and validation data. For the purposes of this assignment,
you should preserve this partition. Also, you should only use the
HLA-DRB1*15:01 molecule (look for files containing "1501").
Each row in the CSV files contains two columns: a string representing
the sequence of amino acids and a measure of binding affinity. Here
is an example:
DLDKKETVWHLEE,0
DKSKPKVYQWFDL,0
HYTVDKSKPKVYQ,0
KSKPKVYQWFDLR,0
LHYTVDKSKPKVY,0
TVDKSKPKVYQWF,0
VDKSKPKVYQWFD,0
YTVDKSKPKVYQW,0
ADVILPIGTRSVETD,0.646144
AFKPVLVDEGRKVAI,0.57075
AGLKTNDRKWCFEGP,0.615622
AHLAEENEGDNACKR,0
ALYEKKLALYLLLAL,0.610019
ANGKTLGEVWKRELN,0.495407
Note that different rows contain different numbers of amino acids.
The binding affinity is encoded on a log scale and ranges between 0
and 1. Peptides with affinities at or above 0.426 are considered
more likely than not to bind to the molecule in question, whereas
affinities below this threshold are considered less likely to bind.
Deep Learning Experiment
Objective: Create a recurrent neural network that predicts binding
affinity as a function of the amino acid string.
- You will need to load the CSV files and create the
tensorflow-compatible data sets. I suggest that you:
- Use a character-based encoding layer for the strings
(this is effectively a 1-hot encoding for each of the
characters in the string without the overhead of so many
zeros; see the RNN text preprocessing section in the
book)
- "Zero-pad" the strings that are short with a special
character
- Follow the encoding layer with one or more trainable embedding
layer(s).
- Use your favorite RNN module
- Use return_sequences=False
- Use one or more dense layers to predict the affinity measure.
Think about what the appropriate non-linearity is for the
output unit
Performance Reporting
Once you have selected a reasonable architecture and set of
hyper-parameters, produce the following figures:
- AUC as a function of epoch for each of the training folds (so,
5 curves)
- AUC as a function of epoch for each of the validation folds
- Accuracy as a function of epoch for each of the training
folds. Note: using tf.keras.metrics.BinaryAccuracy() as a
metric will allow you to set the threshold of 0.426
- Accuracy as a function of epoch for each of the validation folds
In addition, report the following:
- Report the average AUC (across the folds) for the training,
validation and test sets. An average validation AUC of 0.8 is doing
well
- Report the average accuracy (across the folds) for the training,
validation and test sets. An average validation accuracy of 0.84 is
doing well
Provided Code
In the git repository:
- hla_support.py: prepare_data_set() will load and prepare a data set for training/validation/testing
- The "ins" are token indices (not a 1-hot encoding).
- This function splits the training data set into a
training and validation data set. Leave the seed and
the size at their default values (this way, we can
better compare our results)
- Hyper-parameter searches should only be done using the
performance of the validation data set
- You can evaluate the performance of the model on the
test set using:
model.evaluate(ins_test, outs_test)
BUT: only look at these values after you have selected your favorite hyper-parameters
- Use
a tf.keras.layers.Embedding as the first layer of
your network - it will properly handle the token indices
- metrics_binarized.py: Useful metrics
- The "outs" are log affinity measures. However, any
peptide with an affinity at or above 0.426 is considered
as "bindable" to the allele in question.
- You may formulate your problem entirely in terms of a
binary prediction problem, in which case you will
convert the "outs" to binary vectors and use
cross-entropy as your loss
- You may also choose to predict the log affinity
directly, in which case you might use mse as your loss.
However, in order to report Accuracy or AUC, you will
need to binarize the true values of the outs. The
provided wrapper classes do just this for you
What to Hand In
Hand in a PDF file that contains:
- Code for generating and training the network. Some useful UNIX
command line programs:
- enscript: translate code (e.g., py files) into postscript files
- ps2pdf: translate postscript files into pdf files
- pdfunite: merge several pdf files together
- The above figures
- The final metrics (note that this can be with respect to the
best performing epoch, as identified by EarlyStopping)
Grades
- 50 pts: Model generation code. Is it correct? clean? documented?
- 50 pts: Model figures and performance.
- 10 pts: An average validation AUC of 0.82 or better
References
andrewhfagg -- gmail.com
Last modified: Wed Apr 7 21:54:42 2021

